Oligo Datasets
The oligo datasets currently host oligo sequences of 9 species, including six plant species (Arabidopsis, rice, maize, potato, barley and soybean), two mammals (human and mouse) and a model species zebrafish. The details of the oligos are showed in the bellow table.
To download the oligos, please click the download link of the speices. Details about Chorus2, which was used to design the oligos can be found here.
Information table
| Species | Scientific name | Reference | No. of oligos | Download |
|---|---|---|---|---|
| Arabidopsis | Arabidopsis thaliana | TAIR10 | 1,089,367 | bed |
| Rice | Oryza sativa | TIGR7 | 1,710,279 | bed |
| Maize | Zea mays | B73_v4 | 1,641,770 | bed |
| Potato | Solanum tuberosum | DM_v4.04 | 1,674,902 | bed |
| Barley | Hordeum vulgare | IBSC_v2 | 2,968,560 | bed |
| Soybean | Glycine max | Gmax_ZH13_v2.0 | 1,851,420 | bed |
| Human | Homo sapiens | hg38 | 2,311,370 | bed |
| Mouse | Mus musculus | mm10 | 3,397,774 | bed |
| Zebrafish | Danio rerio | danRer11 | 2,442,308 | bed |
Images
 |
 |
 |
| Arabidopsis | Rice | Maize |
 |
 |
 |
| Potato | Barley | Soybean |
 |
 |
 |
| Human | Mouse | Zebrafish |
How to use the datasets
To use the oligo sequences of the target species, users should first download the bed file from the download column in the above table.
Oligo sequences are provided with bed.bgz format, which is a compressed version of bed file.
Users can decompress the file following the below instructions:
For Windows Users:
Download 7-Zip software and install.
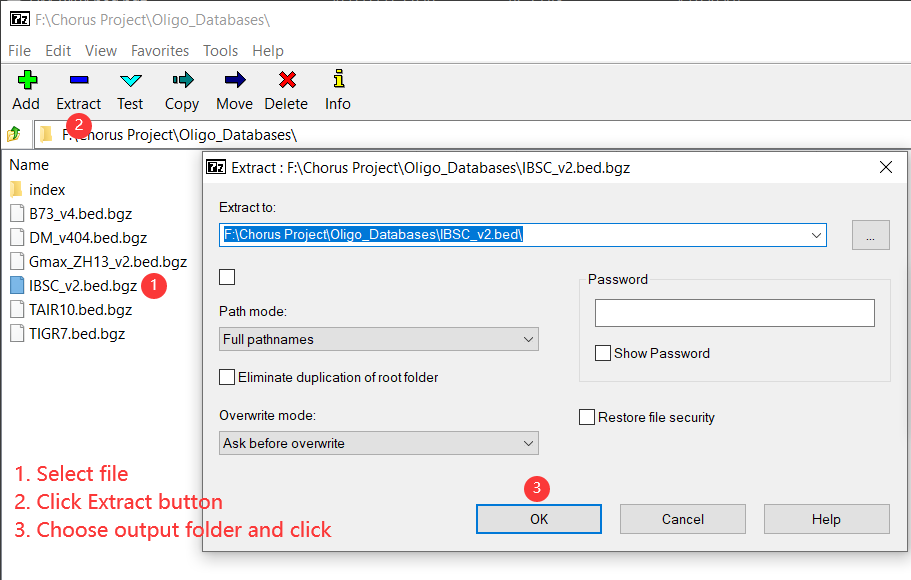
Use 7-Zip to uncompress bed.bgz file.
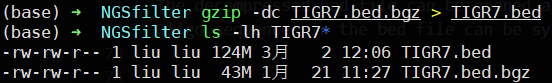
For Linux/MacOS Users:
Using the following command to uncompress bed.bgz file:
$ gzip -cd xxx.bed.bgz > xxx.bed.
The decompressed bed file can be opened and read by text editor or Excel easily.
For Windows users, text editor (Such as EditPlus) is the optimum choice to open it.
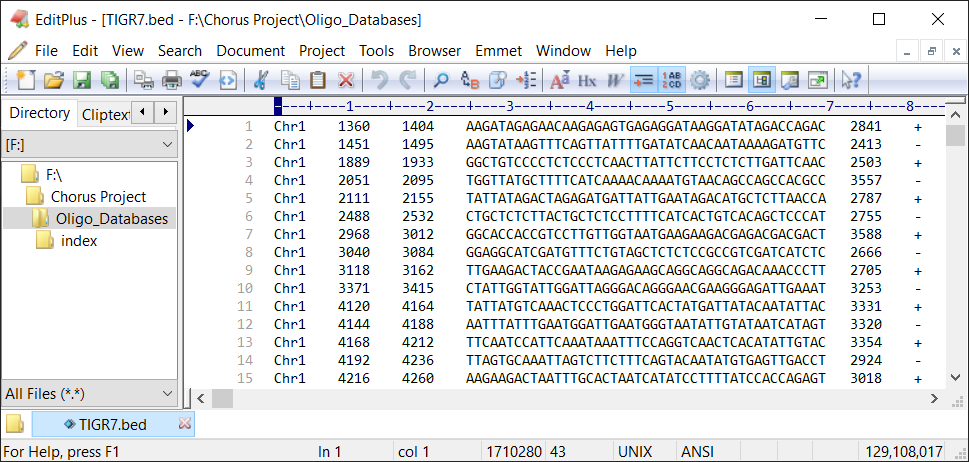
The bed file contains six columns which are separated by delimiter, just like this:
Chr1 1360 1404 AAGATAGAGAACAAGAGAGTGAGAGGATAAGGATATAGACCAGAC 2841 +
Each column represents chromosome, oligo start site, oligo end site, oligo probe sequence, k-mer score and target strand of probes, respectively.
Windows Users can use the filter function of Excel to select the target oligos.
For Linux/MacOS users, awk or perl command may be a better method to select the desired oligo sequences. Just like this:
awk '$1=="Chr1"&&$2>=100000&&$3<=200000' TIGR7.bed
This command will extract oligos in the region Chr1:100000-200000 in rice.

Finally, oligo sequences in the fourth column of the bed file can be synthesized directly for oligo-FISH experiments.
Chorus2
Chorus2 is a software which is developed to design genome-scale oligonucleotide-based probes for fluorescence in situ hybridization (FISH).
Chorus2 uses python script Chorus2.py to identify and pre-filter oligos. It is implemented with a “k-mer score” method to remove repetitive oligos of target genome. Chorus2 run fast and can handle large genome like wheat. The oligos designed by Chorus2 has high specificity and suitable for FISH. The Chorus2 package runs on Linux, macOS and Windows with flexible command-line or an easy-to-use GUI (graphical user interface).
* If there are any questions when using Chorus2 or our oligo datasets, please contact us (hanyangshuo@zhangtaolab.org or liuguanqing@zhangtaolab.org).